ClinicalAI
Problem & Motivation
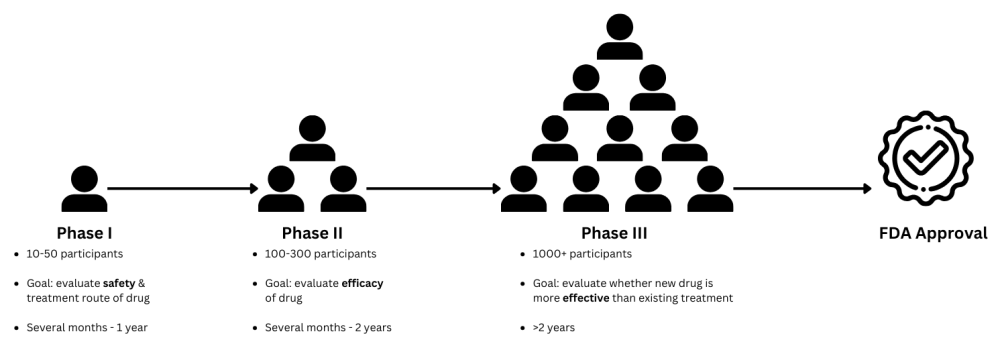
Within the pharmaceutical industry, clinical trials are essential for the delivery of new drugs or treatment to the public. At minimum, a novel drug has to undergo 3 phases of study in order to receive FDA approval. Phase 3 trials are usually the last and most difficult hurdle to clear due to the scale of their complexity and duration, taking anywhere between 2 to 15 years.
Because of their significant logistical complexity and purpose, Phase 3 trials have a very high risk of both delays and failure, which directly corresponds to delays in getting life-saving drugs to patients. In addition, the success of trials is typically dependent on the personal experience of the trial organizer. We hypothesize that providing any kind of non-subjective and non-empirical insight for trial duration has the potential to increase chances of trial success.
Despite numerous trials and a global market valuation of $55.86 billion in clinical research services, there lacks a robust, publicly accessible tool for predicting trial durations. Our work aims to fill this gap by developing a machine learning pipeline that provides duration estimates for Phase 3 oncology trials, the most resource-intensive phase in drug development.
We propose a Clinical AI Web API that uses machine learning to predict the duration of clinical trials. This tool aims to serve Clinical Research Organizations (CROs), pharmaceutical companies, and researchers. By entering their preliminary study parameters, the API will provide them with accurate predictions that can aid in better resource management, budgeting, and strategic planning.
Data Source & Data Science Approach
Our data is from ClinicalTrials.gov, a prominent global and open source registry for clinical research studies. Our project utilizes 19,049 oncology studies:
- 5,053 Phase 1 studies
- 5,982 Phase 2 studies
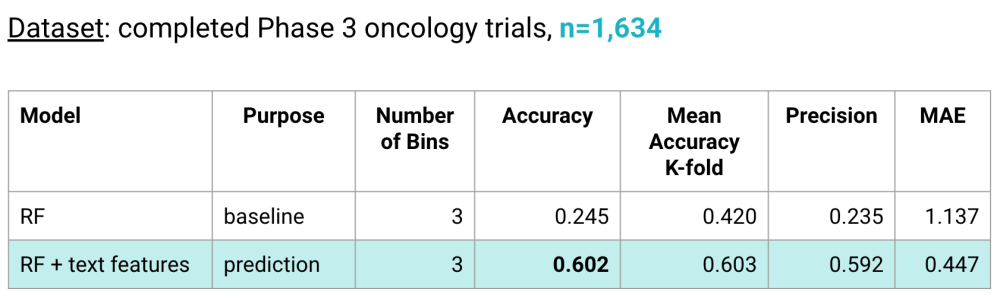
- 1,634 Phase 3 studies
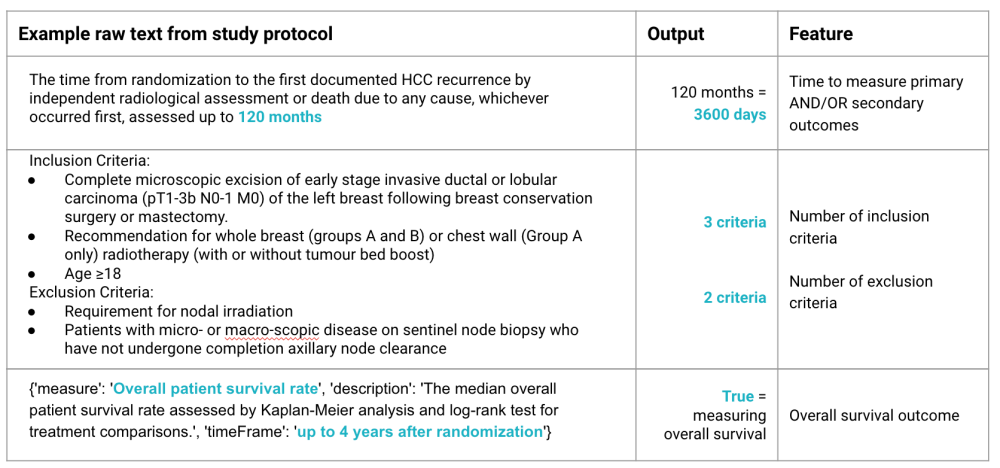
Study protocol data includes information on study locations, number of patients enrolled, disease types, and outcome measurement methods. We were able to extract novel features relevant to trial duration from the text heavy data fields of study protocols through a combination of looking for string and regex patterns. These text fields are highly variable in structure and formatting because they are human-written. Extracted features included (but were not limited to):
- Cycle time to measure patient treatment outcomes
- Number of inclusion and exclusion criteria for study participants
- Whether the trial measures overall survival of the participant
See the table below for examples of raw text and the extracted feature:
The importance of these novel features was confirmed through both feature importance ranking and the performance of our prediction model.
Our prediction model is a Random Forest (RF) classifier trained with our novel extracted text features that predicts a time interval for Phase 3 trial duration. Our baseline was also a RF classifier that was trained only on the most basic study protocol features, i.e. features that did not require extraction and could be taken directly from the study protocol. We observed a +0.4 improvement in accuracy of our prediction model over the baseline.
Evaluation
In addition to predicting on a test and k-fold cross validation, we evaluated our prediction model with a feature ablation study. We found that some of our highest ranked features were actually correlated with one another, e.g. number of inclusion criteria was correlated with number of exclusion criteria, time to measure primary treatment outcomes was correlated with time to measure secondary treatment outcomes. When one of the correlated feature pairs was removed from the model, performance in terms of accuracy and mean absolute error did not suffer since the other feature was still in the model. This means in future steps, we should be able to reduce the number of features users will need to enter into the MVP.
Key Learnings & Impact
- We learned study protocol data alone is not sufficient for high accuracy predictions of trial duration.
Our original intention was to use regression models to predict trial duration rather than a binned time interval. However, we found that this dataset has very high variance in terms of trial duration due to the nature of oncology studies, e.g. adverse events, such as patient death, would extend recruitment and trial times. We attempted to take into account the possible frequency of adverse events by mapping survival rates to different cancer types.
- Using large language models (LLMs) for feature extraction is difficult and a nontrivial task with jargon-heavy text.
We have shown the significance of extracting features from protocol text, but our current methods are with hard coding. We did attempt to use LLMs for feature extraction, but experienced considerable difficulty with getting consistent and accurate extractions. We theorize this is maybe because of the specific medical jargon being used. Although we attempted to use a Named Entity Recognition (NER) model trained on biomedical text, we were still unable to get good extractions. Further time and investment is required to explore using LLMs for feature extraction in this context.
- Our model trained on Phase I data shows promise for outperforming the current best published duration prediction model.
Lastly, we also trained models on Phase I and Phase II oncology data and found that our Phase I model with 2 bins shows promise for outperforming the current best published clinical trial duration prediction model. Future work includes further evaluating the Phase I model with the intent of publication.
Although we weren’t able to achieve the model accuracy we were hoping, we were still able to expand on existing knowledge in this problem space and this work has still served our greater mission: to improve the quality and efficiency of clinical trials to better deliver novel therapeutic solutions to patients in need.
Acknowledgements
We thank our Capstone instructors, Puya Vahabi and Korin Reid, for their technical guidance and feedback throughout the course.
We thank Stephanie Wong for her contributions to the UX design of our MVP and branding.